New research led by scientists at Arizona State University has revealed some of the first detailed molecular clues associated with one of the leading causes of death and disability, a condition known as traumatic brain injury (TBI).
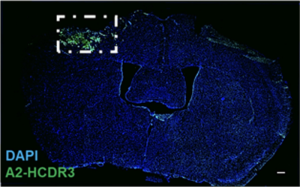
Qualitative representation of A2 acute injury-specific TBI biomarker HCDR3 (green) and cell nuclei (blue) in 1-dpi tissue. Region of interest (ROI) represented in white box. Scale bars, 200 μm. Image courtesy Sarah Stabenfeldt/ASU
TBI is a growing public health concern, affecting more than 1.7 million Americans at an estimated annual cost of $76.5 billion dollars. It is a leading cause of death and disability for children and young adults in industrialized countries, and people who experience TBI are more likely to develop severe, long-term cognitive and behavioral deficits. Qualitative representation of A2 acute injury-specific TBI biomarker HCDR3 (green) and cell nuclei (blue) in 1-dpi tissue. Region of interest (ROI) represented in white box. Scale bars, 200 μm. Image courtesy Sarah
“Unfortunately, the molecular and cellular mechanisms of TBI injury progression are multifaceted and have yet to be fully elucidated,” said Sarah Stabenfeldt, an ASU associate professor and the leader and corresponding author of the study, which appears in the journal Science Advances. “Consequently, this complexity affects the development of diagnostic and treatment options for TBI; the goal of our research was to address these current limitations.”
Their research approach was to perform a “biopanning” search to reveal several key molecular signatures, called biomarkers, identified directly, immediately after the injury event (the acute phase), and also the long-term consequences (the chronic phase) of TBI.
“For TBI, the pathology evolves and changes over time, meaning that a single protein or receptor may be upregulated at one phase of the injury, but not two weeks later,” Stabenfeldt said. “This dynamic environment makes developing a successful targeting strategy complicated.”
To overcome these limitations, the ASU scientists utilized a mouse model for their study to uncover the root causes of TBI by identifying biomarkers — unique molecular fingerprints found with a given injury or disease.
“The neurotrauma research community is a well-established field that has developed and characterized preclinical animal models to better understand TBI pathology and assess the efficacy of therapeutic interventions,” Stabenfeldt said. “Using the established mouse model enabled us to conduct biomarker discovery where the complexity and evolution of the injury pathology was progressing.”
Scientists can often begin to design therapeutic agents or diagnostic devices based on biomarker discovery. Stabenfeldt’s team used a “bottom up” approach to biomarker discovery.
“Top-down” discovery methods are focused on assessing candidate biomarkers based on their known involvement in the condition of interest,” said study first author Briana Martinez, a recent PhD graduate in Stabenfeldt’s lab. “In contrast, a ‘bottom-up’ method analyzes changes in tissue composition and finds a way to connect those changes to the condition. It’s a more unbiased approach but can be risky because you can possibly identify markers that are not specific to the condition or pathology of interest.”
Next, they utilized several state-of-the-art “biopanning” tools and techniques to identify and capture molecules, including a “bait” technique for fishing out potential target molecules called a phage-display system, in addition to high-speed DNA sequencing to identify protein targets within the genome, and mass-spectrometers to sequence the peptide fragments from the phase display experiments.
One further roadblock to discovery is the unique physiology of a mesh-like network designed to protect the brain from injury or harmful chemicals, called the blood-brain barrier (BBB).
“The blood-brain barrier is a barrier between the vascular and brain tissue,” Stabenfeldt said. “In a healthy individual, the BBB tightly regulates nutrient and waste exchange from the blood to the brain and vice versa, essentially compartmentalizing the brain/central nervous system.
“However, this barrier also complicates drug delivery to the brain so that most molecules/drugs do not passively cross this barrier; therefore, the drug delivery field has sought out ways to modulate both entry and delivery mechanisms. Similarly, for blood-based biomarkers for TBI or other neurodegenerative diseases, specificity to the pathology and transfer of the molecule (if it originates in the brain) from the brain to blood is a challenge.”
When a TBI occurs, the initial injury can disrupt the BBB, which triggers a cascade of cell death, torn, disrupted tissues and debris.
The long-term injury causes inflammation and swelling, and results in the immune response springing into action, but also can lead to an impairment of the brain’s energy sources or can choke off the brain’s blood supply, leading to more neuronal cell death and permanent disability.
A key advantage of their suite of experimental tools and techniques of the phage-display system is that the molecules and potential biomarkers identified are small enough to slip through the tiny holes within the meshwork of the BBB — thus, opening the way to therapeutics based on these molecules.
So, despite all these obstacles, the team found a way.
“Our study leverages the sensitivity and specificity of phage to discover novel targeting motifs,” Stabenfeldt said. “The combination of phage and NGS (next-generation sequencing) has been used previously, thereby leveraging bioinformatic analysis. The unique contribution of our study is putting all of these tools together specifically for an in vivo model of TBI.”
They found a suite of unique biomarkers associated with only the acute or chronic phases of TBI. In the acute phase, TBI targeting motif recognized targets associated with mainly metabolic and mitochondrial (the powerhouse of the cell) dysfunction, whereas the chronic TBI motif was largely associated with neurodegenerative processes.
“Our method for biomarker discovery was sensitive enough to detect injury in brains that were collected at different points in the experiments,” Martinez said. “It was really interesting to see that proteins involved in neurodegenerative diseases were detected at seven days post-injury, but not at the earlier, one-day post-injury timepoint. The fact that we were able to observe these differences really showcases how useful this method could be in exploring various aspects of brain injury.”
It may also begin to explain why people who have had a TBI are more susceptible to developing neurodegenerative diseases like Parkinson’s and Alzheimer’s later in life.
This successful discovery pipeline will now serve as the foundation for the next-generation targeted TBI therapeutics and diagnostics.
Next, the group plans to further its collaborations with ASU’s clinical partners and expand their studies to begin to look for these same molecules in human samples.
The publication also includes key contributions from a team of ASU scientists, including Gergey Alzaem Mousa, Kiera Fleck, Tara MacCulloch, Chris W. Diehnelt and Nicholas Stephanopoulos. ASU departments and schools involved were the School of Biological and Health Systems Engineering in the Ira A. Fulton Schools of Engineering, School of Life Sciences, School of Molecular Sciences, Biodesign Institute Center for Molecular Design and Biomimetics, and Biodesign Institute Center for Innovations in Medicine.
SOURCE: https://news.asu.edu/20220726-asu-scientists-find-molecular-clues-behind-traumatic-brain-injury
