Through a process known as strand displacement, a tiny walking device composed of DNA moves across a surface in a cartwheeling motion. The new device performed this feat more rapidly than any DNA walker designed to date.

Hao Yan is the Milton D. Glick Distinguished Professor of Chemistry and Biochemistry at ASU and director of the Biodesign Center for Molecular Design and Biomimetics. (Photo: ASU Biodesign Institute)
Dynamic DNA nanotechnology often makes use of toehold-mediated strand displacement reactions. In this example, the red strand binds to the single stranded toehold region on the green strand (region 1), and then in a branch migration process across region 2, the blue strand is displaced and freed from the complex. Reactions like these are used to dynamically reconfigure or assemble nucleic acid nanostructures. In addition, the red and blue strands can be used as signals in a molecular logic gate.
When it comes to matching simplicity with staggering creative potential, DNA may hold the prize. Built from an alphabet of just four nucleic acids, DNA provides the floorplan from which all earthly life is constructed.
But DNA’s remarkable versatility doesn’t end there. Researchers have managed to coax segments of DNA into performing a host of useful tricks.
DNA sequences can form logical circuits for nanoelectronic applications. They have been used to perform sophisticated mathematical computations, like finding the optimal path between multiple cities. And DNA is the basis for a new breed of tiny robots and nanomachines. Measuring thousands of times smaller than a bacterium, such devices can carry out a multitude of tasks.
In new research, Hao Yan of Arizona State University and his colleagues describe an innovative DNA walker, capable of rapidly traversing a prepared track. Rather than slow, tentative steps across a surface, the DNA acrobat cartwheels head over heels, covering ground 10- to 100-fold faster than previous devices.
“It is exciting to see that DNA walkers can increase their speed significantly by optimizing DNA strand length and sequences, the collaborative effort really made this happen,” Yan said.
Yan is the Milton D. Glick Distinguished Professor of Chemistry and Biochemistry at ASU and director of the Biodesign Center for Molecular Design and Biomimetics.
The study was led by Nils G. Walter, Francis S. Collins Collegiate Professor of Chemistry, Biophysics & Biological Chemistry, founding director of the Single Molecule Analysis in Real-Time (SMART) Center and founding co-director of the Center for RNA Biomedicine at the University of Michigan, and his team, along with collaborators from the Wyss Institute, the Dana Farber Cancer Institute and the Department of Biological Chemistry at Harvard (all in Boston, Massachusetts).
“The trick was to make the walker go head over heels, which is so much faster than the hopping used before – just as you would see in a kung fu action movie where the hero speeds up by cartwheeling to catch the villain,” says Walter.
The improvements in speed and locomotion displayed by the new walker should encourage further innovations in the field of DNA nanotechnology.
The group’s findings appear in the advanced online issue of the journal Nature Nanotechnology.
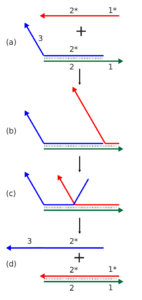
Dynamic DNA nanotechnology often makes use of toehold-mediated strand displacement reactions. In this example, the red strand binds to the single stranded toehold region on the green strand (region 1), and then in a branch migration process across region 2, the blue strand is displaced and freed from the complex. Reactions like these are used to dynamically reconfigure or assemble nucleic acid nanostructures. In addition, the red and blue strands can be used as signals in a molecular logic gate. Credit: Antony-22 Public domain
Building with DNA
Nanoarchitects build their DNA structures, motors and circuits using the same basic principle as Nature. The four nucleotides, labeled A,T,C and G, bind to each other according to a simple and predictable rule: Cs always pair with Gs and As always pair with Ts. Thus, varying lengths of DNA may be programmed to self-assemble, snapping together to form an unlimited variety of two- and 3-dimensional nanostructures. With clever refinement, researchers have been able to outfit their once-static nano-creations with dynamical properties.
One of the more innovative applications of DNA nanotechnology has been the design of robotic walking devices composed of DNA strands that successively move in a stepwise fashion across a path. The method enabling DNA segments to stroll across a defined area is known as strand displacement.
The process works like this: One leg of the robotic device is DNA strand 1, which is bound to complementary strand 2, through normal base pairing. Strand 1 contains an additional, unpaired sequence dangling from its end, which is known as the toehold.
Next, DNA strand 3 is encountered. This strand is complementary to DNA strand 1 and includes a toehold sequence complementary to DNA strand 1. Once the toehold of strand 3 binds with the toehold of strand 1, it begins sequentially displacing each strand 2 nucleotide, one by one, until strand 2 has been is completely replaced by strand 3. Strand 2 then dissociates from strand 1 and the process can begin again. (See figure 1).
Toehold-mediated strand displacement, which forms the basis of other DNA nanodevices, allows DNA structures to move from one complementary foothold on the walking surface to the next. As each DNA strand is displaced by a new strand, the nano-creature takes a step forward.
Race walking
Successful DNA walkers of various kinds have been designed and have demonstrated the ability to ferry nano-sized cargo from place to place. Until now, however, the strand displacement reactions they rely on have been slow, generally requiring several minutes to move a short distance. This is much slower than naturally occurring processes in living systems like protein motors, which can perform feats of dissociation similar to strand displacement in much faster time frames.
While theoretical calculations suggest that individual operations by such nanodevices should occur in seconds or less, in practice, such operations typically require minutes or even hours. (A recently designed cargo-sorting walker for example required 5 minutes for each step, with foothold spacings just 6 nm apart. This speed was on a par with similar strand-displacement walkers.)
In the new study, researchers sought to optimize this process to see how quickly a walker designed with speed in mind could move. The limiting factor in terms of speed did not appear to be the strand displacement process itself, but rather the lack of fine-tuned optimization in the overall walker design.
The team redesigned their walker for maximum speed and used a fluorescent imaging technique known as smFRET (for single-molecule fluorescence resonance imaging transfer) to chart the DNA walker’s progress and evaluate its subtle kinetic properties.
By altering the lengths of toehold sequences and branching migration points, the stepping rate could be keenly optimized, making for a briskly moving nanorobot that left competitors in t

Through a process known as strand displacement, a tiny walking device composed of DNA moves across a surface in a cartwheeling motion. The new device performed this feat more rapidly than any DNA walker designed to date. Graphic by: Zhuoru Li
he dust, boasting stepping rates a full order of magnitude faster than previous DNA walkers.
Freewheeling nanorobot
Part of the robot’s advantage over its competitors is due to its unusual technique of locomotion. Rather than simply stepping from one surface foothold to the next, the acrobatic walker moves head over heels in a cartwheel fashion, while remaining securely bound to at least one foothold at all times.
The stability of the double-stranded sequences anchoring the base of the robot to the track surface, while the free toehold searches out the next complementary sequence, may be one factor improving the walker’s speed. The cartwheeling design also allows strand displacement to sequentially proceed in a direction away from the foothold surface, which improves efficiency.
Once the walker was optimized, super-resolved single particle tracking was used to observe the device’s movement over a 2D surface studded with footholds for the walker, covering a range of up to 2 microns. The best walker optimized in the study was able to search ~43 foothold sites per minute with a stepping distance of ~ 10nm. Strand displacement occurred at rates of about a tenth of a second. Analysis suggests the device can take hundreds of steps without dissociating.
Future steps
While still lagging behind naturally occurring protein reactions, the optimized cartwheeling walker offers a marked advancement in performance, representing an order of magnitude improvement over earlier versions, while not consuming any fuel. Borrowing further insights from natural systems may allow dynamical DNA devices like the walker to accelerate even more in the future by converting chemical energy into directed speed.
The study underlines the opportunities for optimization of a range of DNA nanostructures, considerably enhancing their speed and versatility.
Written by: Richard Harth, ASU Biodesign Institute
